Publications
In preparation
Schulz, Aimee J.; Zhai, Jingjing; AuBuchon-Elder, Taylor; El-Whalid, Mohamed; Hufford, Matthew B.; Johnson, Lynn C.; Kellogg, Elizabeth A.; La, Thuy; Long, Evan; Miller, Zachary R.; Romay, M. Cinta; Seetharam, Arun; Stitzer, Michelle C.; Wrightsmand, Travis; Buckler, Edward S.; Monier, Brandon; Hsu, Sheng Kai (NA) reelGene: a pipeline for robust evaluation through evolutionary leverage of gene models. In preparation doi: NA
Vath, Richard L.; Fernandes, Samuel B.; Monier, Brandon; Głowacka, Katarzyna; Pederson, Taylor L.; Bernacchi, Carl J.; Ferguson, John; Kromdijk, Johannes (NA) Revealing the genetic architecture of Nonphotochemical quenching in sorghum through high-throughput screening of dynamic photoprotection. In preparation doi: NA
Proceedings and Book Chapters
2017
Monier, Brandon; Peta, Vincent; Mensah, Jerry; Bucking, Heike (2017) Inter-and intraspecific fungal diversity in the arbuscular mycorrhizal symbiosis. Mycorrhiza-Function, Diversity, State of the Art doi: https://doi.org/10.1007/978-3-319-53064-2_12
Journal Articles
2023
Khaipho-Burch, Merritt; Ferebee, Taylor; Giri, Anju; Ramstein, Guillaume; Monier, Brandon; Yi, Emily; Romay, M. Cinta; Buckler, Edward S. (2023) Elucidating the patterns of pleiotropy and its biological relevance in maize. PLOS Genetics doi: https://doi.org/10.1371/journal.pgen.1010664
Sun, Xiaopeng; Xiang, Yanli; Dou, Nannan; Zhang, Hui; Pei, Surui; Franco, Arcadio Valdes; Menon, Mitra; Monier, Brandon; Ferebee, Taylor; Liu, Tao; Liu, Sanyang; Gao, Yuchi; Wang, Jubin; Terzaghi, William; Yan, Jianbing; Hearne, Sarah; Li, Lin; Li, Fengand Dai, Mingqiu (2023) The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize. Nature Biotechnology doi: https://doi.org/10.1038/s41587-022-01470-4
2022
Brandon Monier; Terry M. Casstevens; Peter J. Bradbury; Edward S. Buckler (2022) rTASSEL: An R interface to TASSEL for analyzing genomic diversity. Journal of Open Source Software doi: https://doi.org/10.21105/joss.04530
Bradbury, P J; Casstevens, T; Jensen, S E; Johnson, L C; Miller, Z R; Monier, B; Romay, M C; Song, B; Buckler, E S (2022) The Practical Haplotype Graph, a platform for storing and using pangenomes for imputation. Bioinformatics doi: https://doi.org/10.1093/bioinformatics/btac410
Gage, Joseph L; Mali, Sujina; McLoughlin, Fionn; Khaipho-Burch, Merritt; Monier, Brandon; Bailey-Serres, Julia; Vierstra, Richard D; Buckler, Edward S (2022) Variation in upstream open reading frames contributes to allelic diversity in protein abundance. Proceedings of the National Academy of Sciences doi: https://doi.org/10.1073/pnas.2112516119
2021
Ferguson, John N; Fernandes, Samuel B; Monier, Brandon; Miller, Nathan D; Allen, Dylan; Dmitrieva, Anna; Schmuker, Peter; Lozano, Roberto; Valluru, Ravi; Buckler, Edward S; Gore, Michael A; Brown, Patrick J; Spalding, Edgar P; Leakey, Andrew DB (2021) Machine learning-enabled phenotyping for GWAS and TWAS of WUE traits in 869 field-grown sorghum accessions. Plant physiology doi: https://doi.org/10.1093/plphys/kiab346
2020
Gage, Joseph L; Monier, Brandon; Giri, Anju; Buckler, Edward S (2020) Ten years of the maize nested association mapping population: impact, limitations, and future directions. The Plant Cell doi: https://doi.org/10.1105/tpc.19.00951
2019
Monier, Brandon; McDermaid, Adam; Wang, Cankun; Zhao, Jing; Miller, Allison; Fennell, Anne; Ma, Qin (2019) IRIS-EDA: An integrated RNA-Seq interpretation system for gene expression data analysis. PLoS computational biology doi: https://doi.org/10.1371/journal.pcbi.1006792}
McDermaid, Adam; Monier, Brandon; Zhao, Jing; Liu, Bingqiang; Ma, Qin (2019) Interpretation of differential gene expression results of RNA-seq data: review and integration. Briefings in bioinformatics doi: https://doi.org/10.1093/bib/bby067
2018
Monier, Brandon; McDermaid, Adam; Zhao, Jing; Fennell, Anne; Ma, Qin (2018) IRIS-DGE: An integrated RNA-seq data analysis and interpretation system for differential gene expression. bioRxiv doi: https://doi.org/10.1101/283341
McDermaid, Adam; Monier, Brandon; Zhao, Jing; Ma, Qin (2018) ViDGER: An R package for integrative interpretation of differential gene expression results of RNA-seq data. bioRxiv doi: https://doi.org/10.1101/268896
Theses
2018
Monier, Brandon (2018) Microbial communities and their impact on bioenergy crops in dynamic environments. doi: https://openprairie.sdstate.edu/etd/2659
2013
Monier, Brandon (2013) The Analysis of Cytotypic Variation and Construction of a BAC Library of Midwestern Prairie Cordgrass (Spartina pectinata Link) Genotypes. doi: https://openprairie.sdstate.edu/etd/1624
Stats
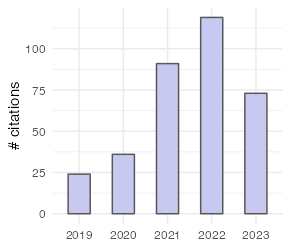
| Metric | All | Since 2018 |
|---|---|---|
| Citations | 357 | 355 |
| h-index | 7 | 7 |
| i10-index | 5 | 5 |
Last Compiled: 2023-07-27